Random forest analysis: Cold-pain tolerance
Peter Kamerman
First version: January 28, 2016
Latest version: September 05, 2016
Session setup
# Load packages
library(ggplot2)
library(scales)
library(grid)
library(cowplot)
## Warning: package 'cowplot' was built under R version 3.2.5
library(readr)
## Warning: package 'readr' was built under R version 3.2.5
library(dplyr)
## Warning: package 'dplyr' was built under R version 3.2.5
library(tidyr)
## Warning: package 'tidyr' was built under R version 3.2.5
library(knitr)
## Warning: package 'knitr' was built under R version 3.2.5
library(party)
## Warning: package 'zoo' was built under R version 3.2.5
# Palette
palette = c('#000000', '#FF0000')
# knitr chunk options
opts_chunk$set(echo = TRUE,
warning = TRUE,
message = FALSE,
cache = TRUE,
fig.path = './figures/cold-pain-tolerance/',
fig.width = 11.69,
fig.height = 8.27,
dev = c('png', 'pdf'),
tidy = FALSE)
Load data
data <- read_csv('./data/random-forest.csv')
Quick look
dim(data)
## [1] 212 14
names(data)
## [1] "ID" "CPT" "PPT" "Race" "Sex"
## [6] "Anxiety" "Depression" "PCS_trait" "PCS_state" "APBQ-F"
## [11] "APBQ-M" "Education" "Assets" "Employment"
head(data)
## # A tibble: 6 × 14
## ID CPT PPT Race Sex Anxiety Depression PCS_trait PCS_state
## <int> <int> <int> <chr> <chr> <dbl> <dbl> <int> <int>
## 1 1 217 874 W M 1.5 1.00 11 10
## 2 2 300 1154 W M 2.4 1.53 15 20
## 3 4 39 741 B F 2.3 1.60 21 11
## 4 5 53 1100 B F 2.3 1.40 23 24
## 5 6 300 1100 B M 1.3 1.27 13 5
## 6 7 300 1249 W M 1.4 1.13 3 2
## # ... with 5 more variables: `APBQ-F` <dbl>, `APBQ-M` <dbl>,
## # Education <int>, Assets <dbl>, Employment <int>
tail(data)
## # A tibble: 6 × 14
## ID CPT PPT Race Sex Anxiety Depression PCS_trait PCS_state
## <int> <int> <int> <chr> <chr> <dbl> <dbl> <int> <int>
## 1 254 111 664 W F 1.6 1.80 12 8
## 2 255 15 524 B F 2.2 2.13 NA NA
## 3 258 300 1126 B M 1.2 1.20 NA 8
## 4 259 70 1117 B F 1.7 2.00 7 10
## 5 260 53 892 B F 1.4 NA 37 12
## 6 262 41 471 B M 1.0 1.27 27 NA
## # ... with 5 more variables: `APBQ-F` <dbl>, `APBQ-M` <dbl>,
## # Education <int>, Assets <dbl>, Employment <int>
Process data
# Clean data
data <- data %>%
mutate(Ancestry = factor(Race),
Sex = factor(Sex),
APBQF = `APBQ-F`,
APBQM = `APBQ-M`,
Education = factor(Education, ordered = TRUE)) %>%
select(-c(ID, PPT, Race, PCS_state, `APBQ-F`, `APBQ-M`))
# Complete X and Y variable dataset
data_complete <- data[complete.cases(data), ]
# Length of full dataset (with NAs)
nrow(data)
## [1] 212
# Length of complete cases dataset
nrow(data_complete)
## [1] 156
glimpse(data_complete)
## Observations: 156
## Variables: 11
## $ CPT <int> 39, 53, 300, 300, 300, 300, 18, 42, 300, 130, 44, 3...
## $ Sex <fctr> F, F, M, M, F, M, M, F, M, M, F, F, F, M, F, F, F,...
## $ Anxiety <dbl> 2.3, 2.3, 1.3, 1.4, 2.3, 1.3, 1.7, 1.5, 1.2, 1.3, 1...
## $ Depression <dbl> 1.60, 1.40, 1.27, 1.13, 2.60, 1.53, 1.40, 1.27, 1.2...
## $ PCS_trait <int> 21, 23, 13, 3, 28, 9, 26, 11, 13, 14, 2, 18, 15, 1,...
## $ Education <ord> 3, 3, 3, 3, 3, 3, 2, 3, 3, 3, 3, 3, 2, 3, 3, 3, 3, ...
## $ Assets <dbl> 1.0, 1.0, 0.6, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1...
## $ Employment <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, ...
## $ Ancestry <fctr> B, B, B, W, W, W, W, W, W, W, W, W, W, W, W, W, B,...
## $ APBQF <dbl> 3.6, 0.9, 3.7, 5.7, 6.0, 4.9, 6.0, 3.4, 4.8, 4.7, 0...
## $ APBQM <dbl> 3.7, 1.5, -3.1, 3.4, 5.2, 3.7, 5.5, 3.0, 2.2, 4.7, ...
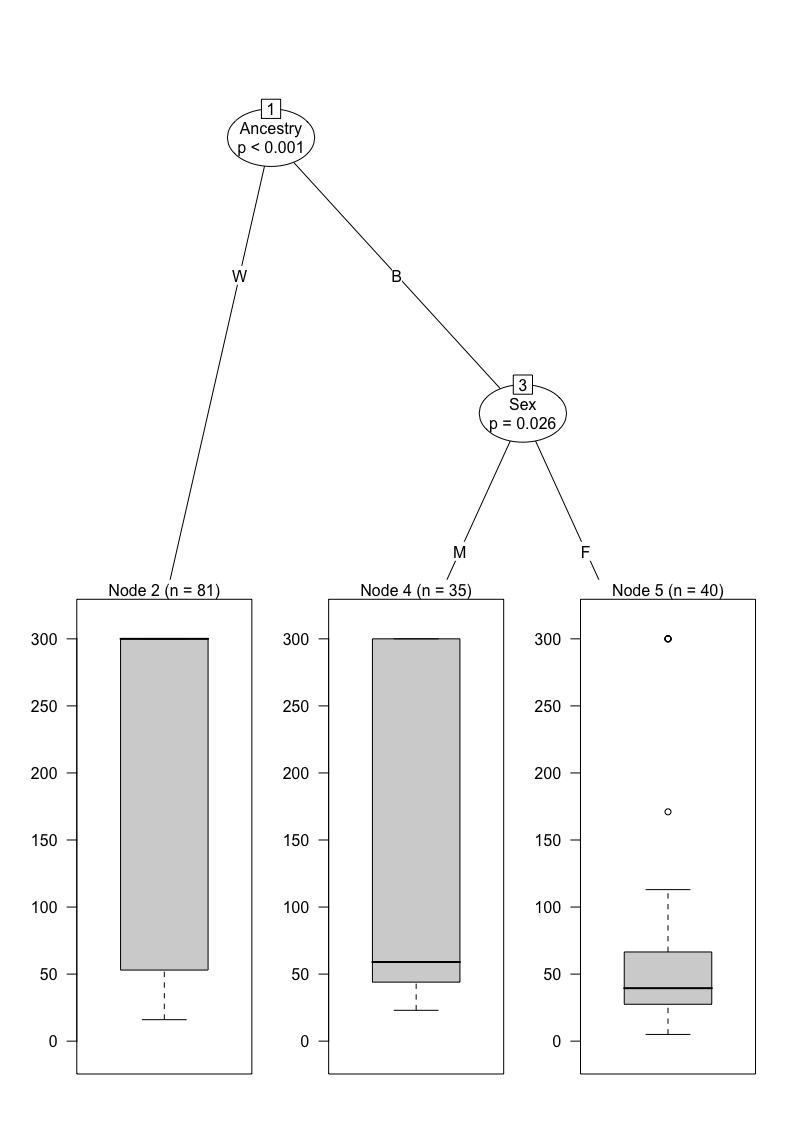
Simple single tree
single_tree <- ctree(CPT ~ ., data = data_complete)
plot(single_tree)
Random Forest
# Set random seeds (used sampling on first run only)
# seed_1 <- sample(1:10000, 1); seed_1
# seed_2 <- sample(1:10000, 1); seed_2
seed_1 <- 816
seed_2 <- 1017
# Data controls
## mtry estimated as sqrt of variables
data.control_1 <- cforest_unbiased(ntree = 500, mtry = 3)
data.control_2 <- cforest_unbiased(ntree = 2000, mtry = 3)
# Model 1
#########
set.seed(seed_1)
# ntree = 500, mtry = 3, seed = seed_1
# Modelling
model_1 <- cforest(CPT ~ .,
data = data_complete,
controls = data.control_1)
model_1_varimp <- varimp(model_1, conditional = TRUE)
# Model 2
#########
# ntree = 2000, mtry = 3, seed = seed_1
# Modelling
model_2 <- cforest(CPT ~ .,
data = data_complete,
controls = data.control_2)
model_2_varimp <- varimp(model_2, conditional = TRUE)
# Model 3
#########
# Set seed
set.seed(seed_2)
# ntree = 500, mtry = 3, seed = seed_2
# Modelling
model_3 <- cforest(CPT ~ .,
data = data_complete,
controls = data.control_1)
model_3_varimp <- varimp(model_3, conditional = TRUE)
# Model 4
#########
# ntree = 2000, mtry = 3, seed = seed_2
# Modelling
model_4 <- cforest(CPT ~ .,
data = data_complete,
controls = data.control_2)
model_4_varimp <- varimp(model_4, conditional = TRUE)
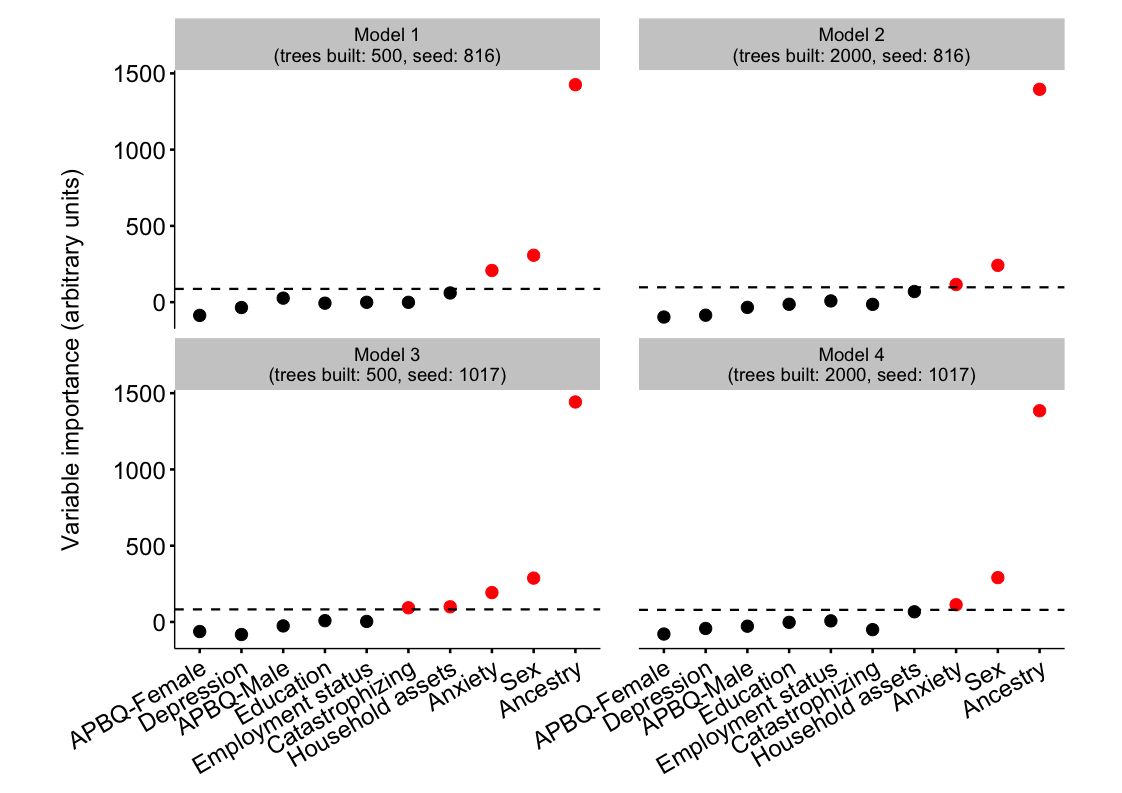
Plots
## Generate plot dataframe
plot_list <- list(model_1_varimp, model_2_varimp,
model_3_varimp, model_4_varimp)
plot_list <- lapply(plot_list, function(x)
data.frame(Variable = names(x), Importance = x, row.names = NULL))
plot_df <- do.call(cbind, plot_list)
plot_df <- plot_df[ , c(1, 2, 4, 6, 8)]
names(plot_df) <- c('Variable', 'Model_1', 'Model_2', 'Model_3', 'Model_4')
plot_df <- plot_df %>%
gather(Model, Value, -Variable) %>%
mutate(Model = factor(Model)) %>%
group_by(Model) %>%
arrange(desc(Value)) %>%
mutate(Important = Value > abs(min(Value)))
## Dataframe of variable importance thresholds
v_importance <- plot_df %>%
summarise(Threshold = abs(min(Value)))
## Vector to label x variables
x_labs <- c(APBQF = 'APBQ-Female',
Depression = 'Depression',
Education = 'Education',
APBQM = 'APBQ-Male',
PCS_trait = 'Catastrophizing',
Assets = 'Household assets',
Anxiety = 'Anxiety',
Sex = 'Sex',
Ancestry = 'Ancestry',
Employment = 'Employment status')
## Vector to order x axis variables
x_order <- plot_df %>%
group_by(Variable) %>%
summarise(mean = mean(Value)) %>%
arrange(mean) %>%
mutate(Variable = factor(Variable, Variable, ordered = TRUE))
x_order <- x_order$Variable
## Vector of facet labels
f_labels <- c(Model_1 = 'Model 1\n(trees built: 500, seed: 816)',
Model_2 = 'Model 2\n(trees built: 2000, seed: 816)',
Model_3 = 'Model 3\n(trees built: 500, seed: 1017)',
Model_4 = 'Model 4\n(trees built: 2000, seed: 1017)')
## Plot
ggplot(data = plot_df, aes(
x = Variable,
y = Value,
colour = Important,
fill = Important)) +
geom_point(size = 4,
shape = 21) +
geom_hline(data = v_importance,
aes(yintercept = Threshold),
linetype = 'dashed',
size = 0.8) +
facet_wrap(~ Model,
labeller = labeller(Model = f_labels)) +
labs(y = 'Variable importance (arbitrary units)\n') +
scale_x_discrete(labels = x_labs,
limits = x_order) +
scale_colour_manual(values = palette) +
scale_fill_manual(values = palette) +
theme(legend.position = 'none',
plot.margin = unit(c(1, 3, 1, 3), 'lines'),
panel.margin.x = unit(2, 'lines'),
axis.title = element_text(size = 18),
axis.title.x = element_blank(),
axis.text = element_text(size = 18),
axis.text.x = element_text(angle = 30, hjust = 1),
axis.line = element_line(size = 0.9),
axis.ticks = element_line(size = 0.9),
strip.text = element_text(size = 14))
Session information
sessionInfo()
## R version 3.2.4 (2016-03-10)
## Platform: x86_64-apple-darwin13.4.0 (64-bit)
## Running under: OS X 10.11.6 (El Capitan)
##
## locale:
## [1] en_GB.UTF-8/en_GB.UTF-8/en_GB.UTF-8/C/en_GB.UTF-8/en_GB.UTF-8
##
## attached base packages:
## [1] stats4 grid stats graphics grDevices utils datasets
## [8] methods base
##
## other attached packages:
## [1] party_1.0-25 strucchange_1.5-1 sandwich_2.3-4
## [4] zoo_1.7-13 modeltools_0.2-21 mvtnorm_1.0-5
## [7] knitr_1.14 tidyr_0.6.0 dplyr_0.5.0
## [10] readr_1.0.0 cowplot_0.6.2 scales_0.4.0
## [13] ggplot2_2.1.0
##
## loaded via a namespace (and not attached):
## [1] Rcpp_0.12.6 formatR_1.4 plyr_1.8.4 tools_3.2.4
## [5] digest_0.6.10 evaluate_0.9 tibble_1.2 gtable_0.2.0
## [9] lattice_0.20-33 Matrix_1.2-6 DBI_0.5 yaml_2.1.13
## [13] coin_1.1-2 stringr_1.1.0 R6_2.1.3 survival_2.39-5
## [17] rmarkdown_1.0 multcomp_1.4-6 TH.data_1.0-7 magrittr_1.5
## [21] codetools_0.2-14 htmltools_0.3.5 splines_3.2.4 MASS_7.3-45
## [25] assertthat_0.1 colorspace_1.2-6 labeling_0.3 stringi_1.1.1
## [29] lazyeval_0.2.0 munsell_0.4.3